Nucleic Acids I: Structure & Replication
Mission 1: Replication
Mission Objectives. You should be able to...
1. Explain how DNA replicates itself.
2. Explain how DNA is packaged.
3. Describe the function of the enzymes involved in replication.
4. Distinguish between the leading strand and lagging strand.
5. State what an Okazaki fragment is and explain its purpose.
6. Analyze the results of the Hershey-Chase experiment.
Mission Objectives. You should be able to...
1. Explain how DNA replicates itself.
2. Explain how DNA is packaged.
3. Describe the function of the enzymes involved in replication.
4. Distinguish between the leading strand and lagging strand.
5. State what an Okazaki fragment is and explain its purpose.
6. Analyze the results of the Hershey-Chase experiment.
DNA Structure
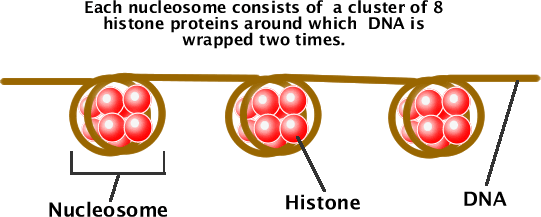
A nucleosome is a basic unit of DNA packaging. It consists of chromatin wrapped around 8 histones, which are positively-charged proteins that adhere to the negatively-charged DNA molecule. The nucleosomes then coil to form solenoids and the loops are then folded to produce a "fiber" that coils once more to form chromatids. This process is called supercoiling and it is how the lengthy DNA molecule is able to fit inside the nucleus. You can read more about supercoiling chromatin here.
The below images show how chromatin coils around the histones to form a nucleosome.
The below images show how chromatin coils around the histones to form a nucleosome.
|
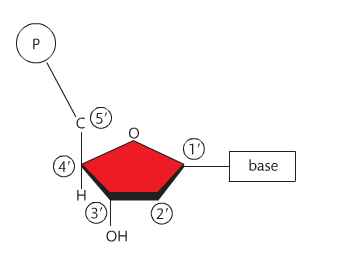
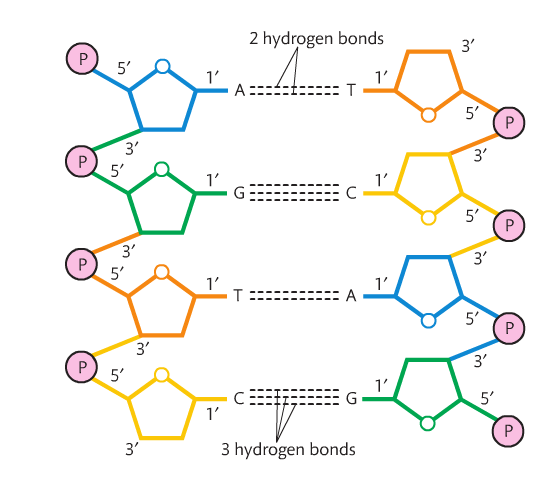
Before we go any further, it is important that you recall the structure of the nucleotide that makes up DNA. It is a 5-carbon sugar with a phosphate and nitrogen base attached. Each carbon has a number. The carbon immediately to the right of the oxygen atom is #1, and goes around the pentagon structure as shown to the right. Carbon #5 is above carbon #4. The nitrogen base is attached to C1 and the phosphate is attached to C5.
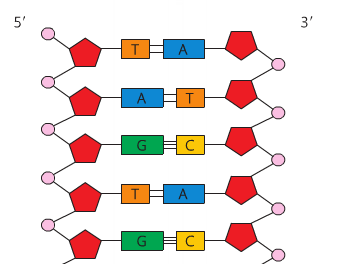
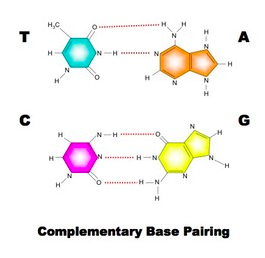
Also recall that the orientation of the sugars change: the 5' carbon has the oxygen at the "top" and the 3' carbon has the oxygen at the bottom. These orientations must be anti-parallel for DNA's double helix structure to work. The structure of DNA includes an alternating sequence of phosphates and deoxyribose sugars. A covalent bond called a phosphodiester bond links the C5 phosphate group to the C3 hydroxyl group. Each nucleotide attaches to another by this method. No matter how many nucleotides are added, there will always be a free 5' carbon with a phosphate group and a 3' carbon with a hydroxyl group. This is the sugar-phosphate backbone of the molecule. As the nucleotides link, a sequence of nitrogen bases becomes clear. The sugar-phosphate backbones are held together by the nitrogen bases and the backbones run in opposite directions (or are antiparallel). So a strand runs 5' - 3' and the opposing strand runs 3' - 5'. There are four nitrogen bases in DNA (adenine, thymine, guanine and cytosine). Adenine forms a double hydrogen bond with thymine and guanine forms a triple hydrogen bond with cytosine. These hydrogen bonds are very weak, which allows for the molecule to be split in replication. Adenine and guanine are known as purines (double ring structures) and guanine and thymine are known as pyrimidines (single ring structures). Single-rings always pair with double-rings which form complementary base pairs based on the distance between the sugar-phosphate backbones. As a result, this dual-stranded molecule forms a double helix when twisted. |
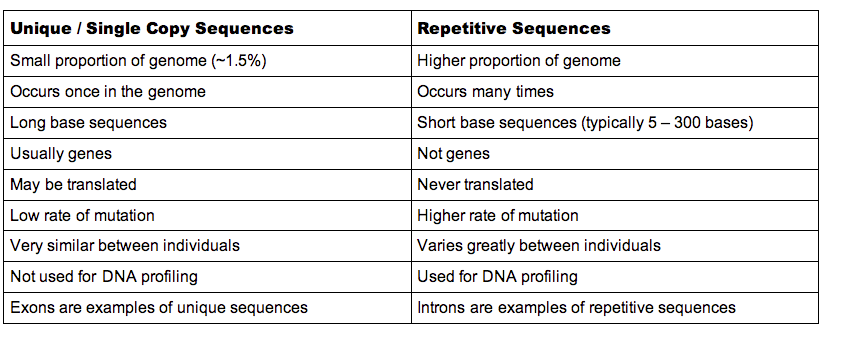
A large portion of the DNA strand is composed of highly repetitive sequences. Then there are the single-copy genes that have coding functions. They are responsible for producing the base sequences that make proteins at the ribosomes, and the base sequences are carried from the nucleus to the ribosomes via messenger RNA. Genes are made up of coding and non-coding fragments called exons and introns, respectively. Structural DNA is DNA that does not have a coding function.
Summary courtesy of Bioninja.
Summary courtesy of Bioninja.
Replication
The complementary base pairing provides a way for DNA to be copied. A single strand provides a template that allows for a perfect copy of the molecule. This is called the semi-conservative model of replication. Replication occurs during the S phase of interphase.
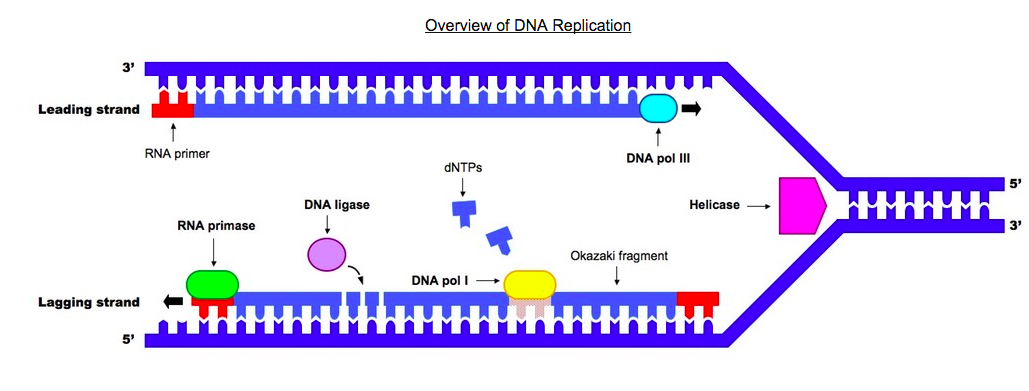
The below image, courtesy of Bioninja, shows how DNA is replicated.
The below image, courtesy of Bioninja, shows how DNA is replicated.
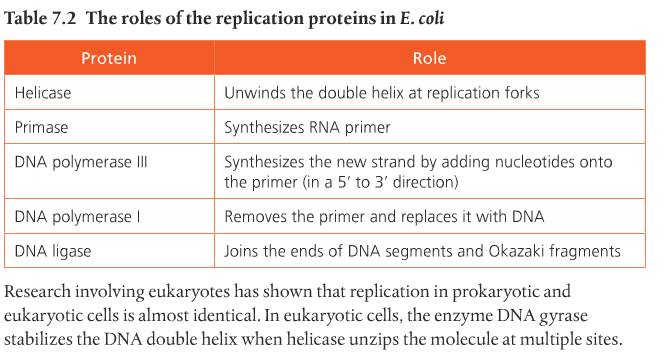
The original strand, shown on the right side of the image, is unzipped or unwound by the enzyme helicase. Helicase breaks the hydrogen bonds that link the nitrogen bases and allows the molecule to split. This occurs at specific regions, called replication origins, that create a replication fork of two polynucleotide strands in antiparallel directions (the middle portion of the diagram). DNA polymerase is an enzyme that helps put together new DNA strands from the original strand, nucleotide by nucleotide.
Remember: Helicase splits the parent strand. DNA polymerase reconstructs two daughter strands.
Each nucleotide that is added to the DNA chain is actually a deoxynucleoside triphosphate dNTP molecule. This molecule contains deoxyribose, a nitrogen base, and three phosphate groups. Two phosphate molecules are lost to provide energy for the chemical bonding of the molecules.
In order for this process to work normally, another enzyme called RNA primase (which is a short strand of RNA), precedes DNA polymerase by preparing the molecule in order for it to receive the nucleotides from DNA polymerase. It is the small portion in red in the above image. Then DNA polymerase III allows the addition of free nucleotides in a 5' to 3' direction to produce the growing strand. DNA can only be formed in the 5' to 3' direction because of polymerase III. In the above image, the leading daughter strand (top) is formed from left to right and the lagging strand is formed from right to left. The lagging strand lags because it's being assembled in reverse and it takes a little more time.
Formation of the lagging strand is assembled by fragments being produce moving away from the replication fork in the 5' to 3' direction. The fragments of the lagging strand are called Okazaki fragments. You can see them in the above image. Primer, primase, and DNA polymerase III are required to begin the formation of each Okazaki fragment of the lagging strand, and to begin the formation of the continuously produced leading strand. They are only needed once for the leading strand. When the Okazaki fragments are assembled, an enzyme called DNA ligase attaches the sugar phosphate backbone of the lagging strand fragments to form a single DNA strand.
The rate at which DNA strands are replicated at a rate of up to 4000 nucleotides per second. This is accomplished by having multiple replication sites on each strand. It is an extremely accurate process; few mutations occur, and cells have repair enzymes that detect and correct mistakes when they do happen.
Remember: Helicase splits the parent strand. DNA polymerase reconstructs two daughter strands.
Each nucleotide that is added to the DNA chain is actually a deoxynucleoside triphosphate dNTP molecule. This molecule contains deoxyribose, a nitrogen base, and three phosphate groups. Two phosphate molecules are lost to provide energy for the chemical bonding of the molecules.
In order for this process to work normally, another enzyme called RNA primase (which is a short strand of RNA), precedes DNA polymerase by preparing the molecule in order for it to receive the nucleotides from DNA polymerase. It is the small portion in red in the above image. Then DNA polymerase III allows the addition of free nucleotides in a 5' to 3' direction to produce the growing strand. DNA can only be formed in the 5' to 3' direction because of polymerase III. In the above image, the leading daughter strand (top) is formed from left to right and the lagging strand is formed from right to left. The lagging strand lags because it's being assembled in reverse and it takes a little more time.
Formation of the lagging strand is assembled by fragments being produce moving away from the replication fork in the 5' to 3' direction. The fragments of the lagging strand are called Okazaki fragments. You can see them in the above image. Primer, primase, and DNA polymerase III are required to begin the formation of each Okazaki fragment of the lagging strand, and to begin the formation of the continuously produced leading strand. They are only needed once for the leading strand. When the Okazaki fragments are assembled, an enzyme called DNA ligase attaches the sugar phosphate backbone of the lagging strand fragments to form a single DNA strand.
The rate at which DNA strands are replicated at a rate of up to 4000 nucleotides per second. This is accomplished by having multiple replication sites on each strand. It is an extremely accurate process; few mutations occur, and cells have repair enzymes that detect and correct mistakes when they do happen.